X-ray Crystallography Services
FastLane™ Structures, Gene-to-Structure & Protein-Ligand co-Crystallization
Our X-ray crystallography services platform handles, on average, over 100 projects annually. Through our work with nearly all large pharmaceutical companies, more than 100 biotech companies, and academic groups, we have gathered extensive experience in all areas of structural biology, including all types of crystallization and X-ray crystallographic problems (more than 500 disclosable X-ray and NMR spectroscopy structures have been deposited to the PDB). Moreover, thanks to the high professional level of our employees and the optimization of our work process, we can provide highly competitive prices! Our publications provide many examples of our contributions.
Please see below for the details of our libraries of drug-target structures and gene-to-structure services. In addition, our structural biology platform overview provides details of our X-ray crystallography services.
Please see below for the details of our libraries of drug-target structures and gene-to-structure services. In addition, our structural biology platform overview provides details of our X-ray crystallography services.
You may also search here for the protein of interest:
Library of verified drug targets that have already been expressed, purified, and crystallized at the company (off-the-shelf structures). Purified proteins are available for the immediate start of a project.
Library of verified drug targets for which expression plasmids and protocols for expression, purification, and crystallization are available at the company.
Gene-to-structure services include cloning, expression, purification, characterization, crystallization, and high resolution X-ray structure determination.
Several proteins from the FastLane™ Premium library can also be purchased for scientific use. Please view our catalog of high-purity crystallization-grade recombinant proteins.
FastLane™ Premium library
The FastLane™ Premium library currently contains drug target proteins ready for co-crystallization with ligands. The library includes an extensive collection of kinases, phosphatases, proteases, epigenetic factors, and other drug targets. Using our high-throughput X-ray crystallography pipeline, the turnaround time for structure determination for these proteins is between 2 to 8 weeks (depending on the protein).
- Kinases
Gene Protein BTK Tyrosine-protein kinase BTK CDK2 Cyclin-dependent kinase 2 CDK5 Cyclin-dependent-like kinase 5 CDK7 Cyclin-dependent-like kinase 7 CK2 Casein kinase II subunit alpha CLK3 Dual specificity protein kinase CLK3 DAPK3 Death-associated protein kinase 3 DYRK1A Dual specificity tyrosine-phosphorylation-regulated kinase 1A EPHA7 Ephrin type-A receptor 7 FGFR1 Fibroblast growth factor receptor 1 FRK Non-receptor tyrosine-protein kinase ITK Tyrosine-protein kinase ITK/TSK KIT Tyrosine-protein kinase, Mast/stem cell growth factor receptor Kit LATS2 Serine/threonine-protein kinase LATS2 MAP2K4 Dual specificity mitogen-activated protein kinase kinase 4 NDR2 Serine/threonine-protein kinase 38-like PFKFB3 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 PIM1 Serine/threonine-protein kinase pim-1 PIM3 Serine/threonine-protein kinase pim-3 PIP4K2A Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha PLK1 Serine/threonine-protein kinase PLK1 PLK4 Serine/threonine-protein kinase PLK4 RET Proto-oncogene tyrosine-protein kinase RIPK3 Receptor-interacting serine/threonine-protein kinase 3 ROCK2 Rho-associated protein kinase 2 RSK1 RPS6KA1 Ribosomal protein S6 kinase alpha-1 RSK2 Ribosomal protein S6 kinase SRPK1 SRSF protein kinase 1 STK10 LOK or Serine/threonine-protein kinase 10 STK17A DRAK1 or Serine/threonine-protein kinase 17A STK17B DRAK2 or Serine/threonine-protein kinase 17B TYK2 Non-receptor tyrosine-protein kinase TYK2 Vps34 PIK3C3 Phosphatidylinositol 3-kinase catalytic subunit type 3 VRK1 Serine/threonine-protein kinase VRK1 VRK2 Serine/threonine-protein kinase VRK2 - Epigenetic targets
- Proteases & phosphatases
Gene Protein CathepsinC CPPI, Dipeptidyl peptidase 1 DPP4 Dipeptidyl peptidase 4 Thrombin F2, Human alpha thrombin PTPN1 PTP1B, Tyrosine-protein phosphatase non-receptor type 1 SHP2 PTPN11, Tyrosine-protein phosphatase non-receptor type 11 USP21 UBP21, Ubiquitin carboxyl-terminal hydrolase 21 USP7 UBP7, Ubiquitin carboxyl-terminal hydrolase 7 USP8 UBP8, Ubiquitin carboxyl-terminal hydrolase 8 - Other targets
Gene Protein ALB Human albumin, HSA BCAT2 Branched-chain-amino-acid aminotransferase, mitochondrial BCL2A1 BFL1, Bcl-2-related protein A1 BSSL Bile salt-activated lipase CD11b human Integrin alpha M/CD11b CD28 T-cell-specific surface glycoprotein CD28 CD47 Leukocyte surface antigen CD47 CD137 TNFRSF9, Tumor necrosis factor receptor superfamily member 9 CGAS Cyclic GMP-AMP synthase DHODH Dihydroorotate dehydrogenase EIF4E Eukaryotic translation initiation factor 4E Gal-1 LGALS1, recombinant Galectin-1 Gal-3C LGALS3, recombinant Galectin-3 C-terminal carbohydrate binding domain Gal-8N LGALS8, recombinant Galectin 8 N-terminal carbohydrate binding domain Gal-9N LGALS9, recombinant Galectin 9, N-terminal carbohydrate binding domain Gal-9C LGALS9, recombinant Galectin 9 C-terminal carbohydrate binding domain GART Trifunctional purine biosynthetic protein adenosine-3 (a.a. 808-1010) GMPR2 GMP reductase 2 HER2 Extracellular domain HSP90 Heat shock protein HSP 90-alpha IL-4 recombinant Interleukin-4 IL-13 recombinant Interleukin-13 IL-17A recombinant Interleukin-17A IL-23 IL-23A, Interleukin-23, subunits alpha and beta K-Ras-cys mut recombinant GTPase K-Ras K-RasG12C recombinant GTPase K-Ras G12C K-RasG12D recombinant GTPase K-Ras G12D K-RasG12V recombinant GTPase K-Ras G12C LDHA LDHA, L-lactate dehydrogenase A chain (rabbit) NRP1 Neuropilin-1 PD-1 PDCD1, Programmed cell death 1 PD-L1 PDL1, Programmed cell death 1 ligand 1 PDE4D cAMP-specific 3',5'-cyclic phosphodiesterase 4D PPARgamma Peroxisome proliferator-activated receptor gamma PSD95 DLG4, Disks large homolog 4 RORgamma Nuclear receptor ROR-gamma S100A12 Protein S100-A12 S100A4 Mts1 or Protein S100-A4 S100A9 MRP14, Protein S100-A9 TNF-alpha Tumor necrosis factor TL1A Tumor necrosis factor ligand superfamily member 15 TUBA1A Tubulin alpha-1A (pig) Ubiquitin Ubiquitin-60S ribosomal protein L40 - Non-human proteins
Gene Protein blaC Beta-lactamase (M. tuberculosis) gyrB DNA gyrase subunit B (S. aureus) Snf LeuT, Na(+):neurotransmitter symporter, Snf family (Aquifex aeolicus VF5) SARS CoV-2 RBD Spike glycoprotein receptor-binding domain (SARS-CoV-2) SARS CoV-2 RBD beta Spike glycoprotein receptor-binding domain, B.1.351 variant (SARS-CoV-2)

On the image: Björn Walse, PhD, CEO SARomics Biostructures, and Raymond Kimbung, PhD, Senior Scientist, Team Leader, Protein Production
FastLane™ Standard library
The FastLane™ Standard library currently contains around 500 constructs of drug target proteins ready for expression, purification, crystallization, and X-ray structure determination according to verified protocols. The library includes a large number of protein kinases, phosphatases, epigenetic factors, and many other drug targets. Using our high-throughput X-ray crystallography pipeline, the turnaround time for structure determination for these proteins is between 6 to 12 weeks (depending on the protein).
Please click to view the list of FastLane™ Standard proteins
- Kinases ABL2 - V-abl Abelson murine leukemia viral oncogene homolog 2 (arg, Abelson-related gene) [isoform 1], (a.a. 279-546).
ACVR1 - Activin A receptor, type I (a.a. 201-499).
ACVR2A - Activin A receptor, type IIA (a.a. 191-488).
ACVL1 - Activin A receptor type II-like kinase 1 (a.a. 195-497)
AK1 - Adenylate kinase isoenzyme 1 (a.a. 1-193).
AK2 - Adenylate kinase isoenzyme 2, mitochondrial (a.a. 1-239).
AK4 - Adenylate kinase isoenzyme 4, mitochondrial (a.a. 1-223)
AK5 - Adenylate kinase isoenzyme 5 (a.a. 366-562)
AK3L1 - Adenylate kinase isoenzyme 4, mitochondrial (a.a. 1-223).
AK5 - Adenylate kinase isoenzyme 5 (a.a. 366-562).
AKT3 - v-akt murine thymoma viral oncogene homolog 3 [isoform 1] (a.a. 1-118)
BMPR2 - Bone morphogenetic protein receptor type-2 (a.a. 189-517).
CAMK1G - Calcium/calmodulin-dependent protein kinase type 1G (a.a. 18-316).
CAMK2A - Calcium/calmodulin-dependent protein kinase type II subunit alpha (a.a. 13-302).
CAMK2B - Calcium/calmodulin-dependent protein kinase type II subunit beta (a.a. 11-303).
CAMK1D - Calcium/calmodulin-dependent protein kinase type 1D (a.a. 1-333).
CAMK2D - Calcium/calmodulin-dependent protein kinase type II subunit delta (a.a. 11-309).
CAMK2D - Calcium/calmodulin-dependent protein kinase type II subunit delta (a.a. 5-315)
CAMK2G - Calcium/calmodulin-dependent protein kinase type II subunit gamma (a.a. 5-315).
CAMK2G - Calcium/calmodulin-dependent protein kinase type II subunit gamma (a.a. 5-315)
CAMK4 - Calcium/calmodulin-dependent protein kinase type IV (a.a. 15-340).
CLK1 - Dual specificity protein kinase (a.a. 148-484).
CLK2 - Dual specificity protein kinase (a.a. 135-496).
CSNK1G1 - Casein kinase I isoform gamma-1 (a.a. 45-352).
CSNK1G2 - Casein kinase I isoform gamma-2 (a.a. 43-353).CSNK1G3 - Casein kinase I isoform gamma-3 (a.a. 35-362).
DMPK - Myotonin-protein kinase (a.a. 11-420).
DYRK2 - Dual specificity tyrosine-phosphorylation-regulated kinase 2 (a.a. 146-552).
FES - Tyrosine-protein kinase Fes/Fps (a.a. 448-822)
FLT1 (VEGFR1) - Tyrosine-protein kinase receptor FLT (a.a. 801-1158).
GAK - Cyclin-G-associated kinase (a.a. 12-347).
HASPIN - GSG2 or Serine/threonine-protein kinase haspin (a.a. 465-798).
ITPK1 - Inositol-tetrakisphosphate 1-kinase (a.a. 1-327).
ITPKC - Inositol-trisphosphate 3-kinase C (a.a. 425-683).
LIMK1 - LIM domain kinase 1 [isoform 1], kinase domain (a.a. 330-637)
MAP2K6 - Dual specificity mitogen-activated protein kinase kinase 6 (a.a. 47-334).
MAP3K10 - Mitogen-activated protein kinase kinase kinase 10, SH3 domains (a.a. 13-78)
MAP3K3 - Mitogen-activated protein kinase kinase kinase 3 [isoform 2] PB1 domain (a.a. 27-124).
MAP3K5 - Mitogen-activated protein kinase kinase kinase 5 (MAPK/ERK kinase kinase 5), (a.a. 659-951).
MAPK11 - Mitogen-activated protein kinase 11 (p38b), (a.a. 5-350).
MAPK6 - Mitogen-activated protein kinase 6 (a.a. 9-327).
MAST1 - Microtubule associated serine/threonine kinase 1, PDZ domain (a.a. 965-1057).
MAST3 - Microtubule associated serine/threonine kinase 3, PDZdomain (a.a. 948-1040)
MAST4 - Microtubule-associated serine/threonine-protein kinase 4 , PDZ domain (a.a. 1140-1231)
MPSK1 - STK16 or Serine/threonine-protein kinase 16 (a.a. 13-305).
MST4 - STK26, Serine/threonine-protein kinase (a.a. 1-300).
MYLK4 - Myosin light chain kinase family, member 4 (a.a. 40-388).
NEK2 - Serine/threonine-protein kinase (a.a. 1-271).PCTK1 - CDK16, Cyclin-dependent kinase 16 (a.a. 163-478).
PHKG2 - Phosphorylase b kinase gamma catalytic chain, liver/testis isoform (a.a. 6-293).
PIK3C2G - Phosphatidylinositol 3-kinase C2 domain-containing subunit gamma, PX domain (a.a. 1198-1307).
PIK3C3 - Phosphatidylinositol 3-kinase catalytic subunit type 3 (a.a. 282-879).
PIK3C3 - Phosphatidylinositol 3-kinase catalytic subunit type 3 (a.a. 268-879).
PIK3R2 - Phosphatidylinositol 3-kinase regulatory subunit beta (a.a. 108-298).
PIM2 - Serine/threonine-protein kinase pim-2 (1-311).
PIP5K2C - Phosphatidylinositol 5-phosphate 4-kinase type-2 gamma (a.a. 32-421).
PKM2 - Pyruvate kinase isozymes (a.a. 3-530).
PRKCG - Protein kinase C gamma type, C2 domain (a.a. 154-295).
PTK2B - Protein-tyrosine kinase 2-beta (a.a. 414-692).
ROR2 - Tyrosine-protein kinase transmembrane receptor ROR2 (a.a. 463-751).
RPS6KA1 - Ribosomal protein S6 kinase alpha-1 (a.a. 404-719).
SLK - STE20-like serine/threonine-protein kinase (a.a. 19-320).
SRPK2 - SRSF protein kinase 2 (a.a. 51-688).
TTK - MPS1, Dual specificity protein kinase TTK (a.a. 519-808).
STK16 - Serine/threonine protein kinase 16 (a.a. 13-305).
STK25 - Serine/threonine-protein kinase 25 (a.a. 1-293).
STK26 - Serine/threonine-protein kinase 26 (a.a. 1-300).
TNIK - TRAF2 and NCK interacting protein kinase (a.a. 1-325).
TTK - Dual specificity protein kinase TTK (a.a. 519-808).
VRK3 - Inactive serine/threonine-protein kinase VRK3 (a.a. 146-474).
YSK1 - STK25, Serine/threonine kinase 25 (a.a. 1-293). - Phosphatases CDC25C - M-phase inducer phosphatase 3 (a.a. 270-462)
DUSP13 - Dual specificity protein phosphatase 13 isoform B (a.a. 1-198).
DUSP16 - Dual specificity protein phosphatase 16 (a.a. 5-150)
IMPA2 - Inositol monophosphatase 2 (a.a. 16-288).
INPP5B - Type II inositol 1,4,5-trisphosphate 5-phosphatase (a.a. 339-643)
INPP5E - 72 kDa inositol polyphosphate 5-phosphatase (a.a. 275-623).
INPPL1 - Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 (a.a. 419-732).
ITPA - Inosine triphosphate pyrophosphatase (a.a. 1-194).
LHPP - Phospholysine phosphohistidine inorganic pyrophosphate phosphatase (a.a. 1-270).
MDP1 - Magnesium-dependent phosphatase 1 (a.a. 1-165).
MTMR6 - Myotubularin-related protein 6 (a.a. 1-505).
NANP - N-acylneuraminate-9-phosphatase (a.a. 1-248).
PGAM5 - Serine/threonine-protein phosphatase, mitochondrial (a.a. 90-289)
PHPT1 - 14 kDa phosphohistidine phosphatase (a.a. 5-125).PTPRB - Receptor-type tyrosine-protein phosphatase beta (a.a. 1686-1971).
PPP2R5C - Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform (a.a. 11-380).
PTPA - Serine/threonine-protein phosphatase 2A activator (a.a. 22-358)
PTPN3 - Non-receptor tyrosine-protein phosphatase type 3 (a.a. 628-913).
PTPN4 - Non-receptor tyrosine-protein phosphatase type 4 (a.a. 611-926).
PTPN5 - Non-receptor tyrosine-protein phosphatase type 5 (a.a. 282-563).
PTPN7 - Non-receptor tyrosine-protein phosphatase type 7 (a.a. 65-358).
PTPN9 (MEG2) - Non-receptor tyrosine-protein phosphatase type 9 (a.a. 277-582).
PTPN14 - Non-receptor tyrosine-protein phosphatase type 14 (a.a. 886-1187)
PTPN22 - Non-receptor tyrosine-protein phosphatase type 22 (a.a. 1-302).PTPN18 - Non-receptor tyrosine-protein phosphatase type 18 (a.a. 6-299).
PTPRE - Receptor-type tyrosine-protein phosphatase epsilon (a.a. 107-697).
PTPRG - Receptor-type tyrosine-protein phosphatase epsilon (a.a. 827-1445).
PTPRJ - Receptor-type tyrosine-protein phosphatase eta (a.a. 1019-1311).
PTPRK - Receptor-type tyrosine-protein phosphatase eta (a.a. 865-1154).
PTPRN2 - Receptor-type tyrosine-protein phosphatase N2 (a.a. 715-1010).
PTPRO - Receptor-type tyrosine-protein phosphatase O (a.a. 916-1208).
PTPRR - Receptor-type tyrosine-protein phosphatase R (a.a. 375-655).
PTPRT - Receptor-type tyrosine-protein phosphatase T (a.a. 868-1151).
RNGTT - mRNA-capping enzyme (a.a. 1-219).
THTPA - Thiamine-triphosphatase (a.a. 1-215). - Epigenetic targets: Bromodomains ASH1L - Bromodomain, Histone-lysine N-methyltransferase ASH1L (a.a. 2438-2561).
BACH1 - Transcription regulator protein BACH1, BTB domain (a.a. 7-128)
BAZ2B - Bromodomain adjacent to zinc finger domain protein 2B (a.a. 2054-2168).
BPTF - Bromodomain, Nucleosome-remodeling factor subunit BPTF (a.a. 2914-3037).
BRD1 - Bromodomain-containing protein 1 (a.a. 556-688).
BRD9 - Bromodomain-containing protein 9 (a.a. 134-239).
BRWD1 - Bromodomain and WD repeat-containing protein 1 (a.a. 1310-1430 Bromodomain 2).
CECR2 - Bromodomain, Cat eye syndrome critical region protein 2 (a.a. 424-538).CREBBP - Bromodomain, CREB-binding protein (a.a. 1081-1197).
EP300 - Bromodomain, Histone acetyltransferase p300, (a.a. 1040-1161).
KAT2A - Bromodomain, Histone acetyltransferase KAT2A, (a.a. 729-837).
KAT2B - Bromodomain, Histone acetyltransferase KAT2B (a.a. 715-831).
MINA - Bifunctional lysine-specific demethylase and histidyl-hydroxylase MINA (a.a. 26-465)
PBRM1 - Bromodomain, Protein polybromo-1 (a.a. 43-156).
PBRM1 - Bromodomain, Protein polybromo-1 (a.a. 178-291).
PBRM1 - Bromodomain, Protein polybromo-1 (a.a. 388-494).
PBRM1 - Bromodomain, Protein polybromo-1 (a.a. 645-766). - Epigenetic targets: Demethylases & others JMJD2A - Lysine-specific demethylase 4A (a.a. 1-359).
JMJD2B - KDM4B, Lysine-specific demethylase 4B (a.a. 1-346).
JMJD2C - KDM4C, Lysine-specific demethylase 4C (a.a. 875-995 tudor domains).
JMJD2D - KDM4D, Lysine-specific demethylase 4C (a.a. 12-341).
JMJD3 - Lysine-specific demethylase 6B (a.a.1176-1502).
JMJD6 - Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6 (a.a. 1-335).
KDM4A - Lysine-specific demethylase 4A (a.a. 1-359)
KDM4E - Lysine-specific demethylase 4E (a.a. 1-336)
KDM5C - Lysine-specific demethylase 5C (a.a. 73-188)
KDM6B - Lysine-specific demethylase 6B (a.a. 1176-1505)
MINA - Bifunctional lysine-specific demethylase and histidyl-hydroxylase MINA (a.a. 26-465).
PHF8 - Histone lysine demethylase PHF8 (a.a. 115-483).PARP2 - Poly [ADP-ribose] polymerase 2 (a.a. 235-579).
PARP3 - Poly [ADP-ribose] polymerase 3 (a.a. 178-532).
PARP10 - Poly [ADP-ribose] polymerase 10 (a.a. 09-1017).
PARP12 - Poly [ADP-ribose] polymerase 12(a.a. 89-684).
PARP14 - Protein mono-ADP-ribosyltransferase PARP14 (a.a. 789-979)
PARP14 - Protein mono-ADP-ribosyltransferase PARP14 (a.a. 999-1196)
PARP14 - Poly [ADP-ribose] polymerase 14(a.a. 1611-1801).
PARP15 - Poly [ADP-ribose] polymerase 15 (a.a. 459-656).
TNKS1 - Poly [ADP-ribose] polymerase tankyrase-1 (a.a. 1091-1325)
TNKS2 - Tankyrase-2 (a.a. 946-1162).
TNKS2 - Tankyrase-2 (a.a. 955-1162).
ZC3HAV1 - Zinc finger CCCH-type antiviral protein 1 (a.a. 724-896). - Oxidoreductases ACAD11 - Acyl-CoA dehydrogenase family member 11 (a.a. 353-780)
ACADS - Short-chain specific acyl-CoA dehydrogenase, mitochondrial (a.a. 30-412)
ACADSB - Short/branched chain specific acyl-CoA dehydrogenase, mitochondrial
ACADVL - Very long-chain specific acyl-CoA dehydrogenase, mitochondrial 69-655)
ADH4 - All-trans-retinol dehydrogenase [NAD(+)] ADH4 (a.a. 1-380)
AKR1C4 - Aldo-keto reductase family 1 member C4 (a.a. 1-323)
AKR7A2 - Aflatoxin B1 aldehyde reductase member 2 (a.a. 1-359)
AKR7A3 - Aflatoxin B1 aldehyde reductase member 3 (aa. 6-331)
ALDH7A1 - Alpha-aminoadipic semialdehyde dehydrogenase (a.a. 29-527)
ALDH18A1 - Delta-1-pyrroline-5-carboxylate synthase (362-795)
ASPH - Aspartyl/asparaginyl beta-hydroxylase (a.a. 562-758)
BBOX1 - Gamma-butyrobetaine dioxygenase (a.a. 1-387)
BDH2 - Dehydrogenase/reductase SDR family member 6 (a.a. 1-245)
BLVRA - Biliverdin reductase A (a.a. 7-296)
CBR3 - Carbonyl reductase [NADPH] 3 (a.a. 6-277)
CRYL1 - Lambda-crystallin homolog (a.a. 6-316)
CTBP2 - C-terminal binding protein 2 (a.a. 31-364)
DHRS1 - Dehydrogenase/reductase SDR family member 1 (a.a. 3-262)
DHRS11 - Dehydrogenase/reductase SDR family member 11 (a.a. 1-256)
DHRS4 - Dehydrogenase/reductase SDR family member 4 (a.a. 19-278)
FASN - Fatty acid synthase (a.a. 422-831)
GAPDHS - Glyceraldehyde-3-phosphate dehydrogenase, testis-specific (a.a. 69-407)
GLOD5 - Glyoxalase domain-containing protein 5 (a.a. 34-157)
GLRX2 - Glutaredoxin-2, mitochondrial (a.a. 41-164)
GLRX2 - Glutaredoxin-2, mitochondrial (a.a. 56-164)
GLRX3 - Glutaredoxin-3 (a.a. 1-334)
GLRX3 - Glutaredoxin-3 (a.a. 232-334)
GLRX5 - Glutaredoxin 5
GLYR1 - Cytokine-like nuclear factor N-PAC (a.a. 261-553)
GMPR - GMP reducatse 1 (a.a. 1-345)
GPD1L - Glycerol-3-phosphate dehydrogenase 1-like protein (a.a. 1-349)
GPX1 - Glutathione peroxidase 1 (a.a. 14-198)
GPX2 - Glutathione peroxidase 2 (a.a. 4-188)
GPX3 - Glutathione peroxidase 3 (a.a. 25-223)
GPX4 - Glutathione peroxidase 4 (a.a. 36-197
GPX5 - Epididymal secretory glutathione peroxidase (a.a. 28-220)
GPX7 - Glutathione peroxidase 7 (a.a. 20-177)
GPX8 - Probable glutathione peroxidase 8 (a.a. 44-209)
HAO1 - 2-Hydroxyacid oxidase 1 (a.a. 1-370)
HIBADH - 3-Hydroxyisobutyrate dehydrogenase, mitochondrial (a.a. 41-335)
HPD - 4-hydroxyphenylpyruvate dioxygenase (a.a. 8-393)
HPGD - 15-hydroxyprostaglandin dehydrogenase [NAD(+)] (a.a. 3-256)HSD11B1 - 11-Beta-hydroxysteroid dehydrogenase 1 (a.a. 20-284)
HSD17B10 - 3-hydroxyacyl-CoA dehydrogenase type- 2 (a.a. 1-261)
HSD17B11 - Estradiol 17-beta-dehydrogenase 11 (a.a. 15-275)
HSD17B14 - 17-Beta-hydroxysteroid dehydrogenase 14 (a.a. 1-270)
HSD17B4 - Peroxisomal multifunctional enzyme type 2 (a.a. 1-304)
HSD17B8 - (3R)-3-hydroxyacyl-CoA dehydrogenase Q92506 6-261
GMDS - GDP-mannose 4,6-dehydratase (a.a. 23-372)
MECR - Enoyl-[acyl-carrier-protein] reductase, mitochondrial (a.a. 40-373)
MCEE - Methylmalonyl CoA epimerase (a.a. 45-176)
MDH2 - Malate dehydrogenase 2, NAD (mitochondrial (a.a. 19-338)
ME1 - NADP-dependent malic enzyme (a.a. 13-564)
MSRB1 - Methionine-R-sulfoxide reductase B1 (a.a. 10-112)
NMRAL1 - NmrA-like family domain containing protein 1 (a.a. 1-298)
PECR - Peroxisomal trans-2-enoyl-CoA reductase (a.a. 1-303)
PGD - 6-phosphogluconate dehydrogenase, decarboxylating (1-483)
PHF8- Histone lysine demethylase PHF8 (a.a. 115-483)
PHGDH - D-3-phosphoglycerate dehydrogenase (a.a. 4-314)
PHYHD1 - Phytanoyl-CoA dioxygenase domain-containing protein 1 (a.a. 1-291)
PRDX4 - Peroxiredoxin 4 (a.a. 84-271)
PTGR1 - Prostaglandin reductase 1 Q14914 4-329
PTGR2 - Prostaglandin reductase 2 Q8N8N7 1-350
PTGR3 - Prostaglandin reductase 3 (a.a. 33-371) Q8N4Q0 33-371
PTGR3 - Prostaglandin reductase 3 (a.a. 31-370) Q8N4Q0 31-370
PYCR1 - Pyrroline-5-carboxylate reductase 1, mitochondrial (a.a. 1-300)
RTN4IP1 - Reticulon-4-interacting protein 1, mitochondrial (a.a. 43-396)
SPR - Sepiapterin reductase (a.a. 5-261)
TP53I3 - Quinone oxidoreductase PIG3 (a.a. 1-332)
TXNRD1 - Thioredoxin reductase 1, cytoplasmic (a.a. 150-649)
TXNRD3 - Thioredoxin reductase 3 (a.a. 51-156)
UGDH - UDP-glucose 6-dehydrogenase (a.a. 1-466)
UXS1 - UDP-glucuronic acid decarboxylase 1 (a.a. 85-402) - ATP-ases & GTP-ases ATP-ases
DDX10 - Probable ATP-dependent RNA helicase DDX10 (a.a. 47-280)
DDX18 - ATP-dependent RNA helicase (a.a.149-387)
DDX19B - ATP-dependent RNA helicase (a.a. 54-475)
DDX20 - Probable ATP-dependent RNA helicase (a.a. 41-268)
DDX25 - ATP-dependent RNA helicase (a.a. 307-479)
DDX3X - ATP-dependent RNA helicase (a.a. 168-352)
DDX41 - Probable ATP-dependent RNA helicase (a.a. 402-569)
DDX47 - Probable ATP-dependent RNA helicase (a.a. 5-230)
DDX5 - Probable ATP-dependent RNA helicase (a.a 68-307)
DDX52 - Probable ATP-dependent RNA helicase (a.a. 139-381)
DDX53 - Probable ATP-dependent RNA helicase (a.a 204-430)
DHX9 - ATP-dependent RNA helicase A (a.a. 329-563)
EIF4A1 - Eukaryotic initiation factor 4A-I (a.a. 20-238)
FIGNL1 - Fidgetin-like protein 1, (a.a. 341-674)
HSPA1A - Heat shock 70 kDa protein 1A (a.a. 1-387)
HSPA1L - Heat shock 70 kDa protein 1-like (a.a. 1-386)
HSPA2 - Heat shock-related 70 kDa protein 2 (a.a. 6-386)
HSPA5 - Endoplasmic reticulum chaperone BiP (a.a. 26-410)
HSPA6 - Heat shock 70 kDa protein 6 (a.a. 6-385)
HSPA9 - Stress-70 protein, mitochondrial (a.a. 439-597)
IFIH1 - Interferon-induced helicase C domain-containing protein 1 (a.a. 277-490)
NVL - Nuclear valosin-containing protein-like (a.a. 573-845)
SPG7 - Paraplegin (a.a. 305-565)GTP-ases
ARHGAP15 - Rho GTPase-activating protein 15 (a.a. 262-473)
AGAP2 - Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 2, GTPase like domain (a.a. 402-577)
DIRAS1 - GTP-binding protein Di-Ras1 (a.a. 1-198)
DIRAS2 - GTP-binding protein Di-Ras2 (a.a. 5-176)
GEM - GTP-binding protein GEM (a.a. 62-249)
RAC3 - Ras-related C3 botulinum toxin substrate 3 (a.a. 1-192)
RACGAP1 - Rac GTPase-activating protein 1 (a.a. 348-546)
RHOB - Rho-related GTP-binding protein RhoB (a.a. 4-184)
RHOD - Rho-related GTP-binding protein RhoD (a.a. 7-197)
RHOU - Rho-related GTP-binding protein RhoU (a.a. 32-230)
REM1 - GTP-binding protein REM 1 (a.a. 79-251)
RERG - Ras-related and estrogen-regulated growth inhibitor (a.a. 1-174)
RND1 - Rho-related GTP-binding protein Rho6 (a.a. 5-200)
RRAD - GTP-binding protein RAD (a.a. 87-260)
RRAS - Ras-related protein R-Ras (a.a. 23-201) (a.a. 23-201)
RRAS2 - Ras-related protein R-Ras2 (a.a. 12-181) - Nucleotide metabolism Nucleotide metabolism
ADAT2 - tRNA-specific adenosine deaminase 2 (a.a. 20-185)
ADSL - Adenylosuccinate lyase (a.a. 1-481)
ADSS2 - Adenylosuccinate synthetase isozyme 2 (a.a. 21-456)
CTPS1 - CTP synthase 1, synthase domain (a.a. 1-273)
CTPS2 - CTP synthase 2, glutaminase domain (a.a. 292-562)
CTPS2 - CTP synthase 2, synthase domain (a.a. 1-275)
DCTD - Deoxycytidylate deaminase (a.a. 4-174)
GART - Trifunctional purine biosynthetic protein adenosine-3 (a.a. 1-430)
GART - Trifunctional purine biosynthetic protein adenosine-3 (a.a. 467-794)
GDA - Guanine deaminase (a.a. 1-454)
GMPS GMP synthetase [glutamine hydrolyzing], glutaminase domain (a.a. 11-24,25-219)
GMPS - GMP synthetase [glutamine hydrolyzing] (a.a. 20-693)
ITPKB - Inosine triphosphate pyrophosphatase (a.a. 1-194)
NUDT1 - Oxidized purine nucleoside triphosphate hydrolase (a.a. 1-156)
NUDT6 - Nucleoside diphosphate-linked moiety X motif 6 (a.a. 141-316)
NUDT10 - Diphosphoinositol polyphosphate phosphohydrolase 3-alpha (a.a. 17-144)NUDT14 - Uridine diphosphate glucose pyrophosphatase (a.a. 28-222)
NUDT16 - U8 snoRNA-decapping enzyme (a.a. 1-195)
NUDT16L1 - Protein syndesmos (a.a. 6-211)
NUDT18 - 8-oxo-dGDP phosphatase NUDT18 (a.a. 26-179)
NUDT21 - Cleavage and polyadenylation specificity factor subunit 5 (19-227)
NUDT21 - Cleavage and polyadenylation specificity factor subunit 5 (a.a. 20-227)
PRPSAP1 - Phosphoribosyl pyrophosphate synthase-associated protein 1 (1-351)
PRPSAP2 - Phosphoribosyl pyrophosphate synthase-associated protein 2 (a.a. 12-369)
PRTFDC1 - Phosphoribosyltransferase domain-containing protein 1 (a.a. 1-225)
RRM1 - Ribonucleoside-diphosphate reductase large subunit (a.a. 75-742)
RRM2 Ribonucleoside-diphosphate reductase subunit M2 (a.a. 60-389)
RRM2B - Ribonucleoside-diphosphate reductase subunit M2 B (a.a. 17-322)
UCK1 - Uridine-cytidine kinase 1 (a.a 21-243)
UMPS - Uridine 5'-monophosphate synthase (a.a. 224-479)
UMPS - Uridine 5'-monophosphate synthase (a.a. 7-203)
UPP2 Uridine phosphorylase 2 (a.a.23-317) - Signaling proteins PDZ-domains
DLG2 - Discs, large homolog 2, second PDZ domain (a.a. 418-513)
DLG2 Discs, large homolog 2, first PDZ domain (a.a. 190-283)
DLG3 - Discs, large homolog 3 first PDZ domain (a.a. 126-222)
DLG3 - Discs, large homolog 3 second PDZ domain (a.a. 223-314)
DVL2 - Segment polarity protein dishevelled homolog DVL-2, PDZ domain (a.a. 261.355)
GIPC2 - PDZ domain-containing protein GIPC2 (a.a. 112-200)
GRIP1 - Glutamate receptor interacting protein 1, 2nd PDZ domain (a.a. 148-239)
LNX1 - E3 ubiquitin-protein ligase LNX, PDZ domain (a.a. 504-594)
LNX2 - Ligand of Numb protein X 2, PDZ domain (a.a. 336-425)
MAGI1 - Membrane associated guanylate kinase WW and PDZ containing protein 1, 4th PDZ domain (a.a. 839-922)
MAGI1 - Membrane associated guanylate kinase WW and PDZ containing protein 1, 6th PDZ domain containing C1190S mutant (a.a. 1149-1233)
MAGI1 - Membrane associated guanylate kinase WW and PDZ containing protein 1, 3th PDZ domain continaing double mutant, C677S and C709S (a.a. 640-721)
MPDZ - Multiple PDZ domain protein, 1st PDZ domain (a.a. 117-227)
MPDZ - Multiple PDZ domain protein, 3rd PDZ domain (a.a. 373-463)
MPDZ - Multiple PDZ domain protein, 7th PDZ domain (a.a. 1148-1243)
MPDZ - Multiple PDZ domain protein, 12th PDZ domain (a.a. 1858-1951)
MPDZ - Multiple PDZ domain protein, 13th PDZ domain (a.a. 1983-2070)
MPDZ - Multiple PDZ domain protein, 10th PDZ domain (a.a. 1625-1716)
MPDZ Multiple PDZ domain protein, 11th PDZ domain (a.a. 1722-1806)
NHERF1 - Na(+)/H(+) exchange regulatory cofactor NHE-RF1, PDZ domain (a.a. 150-235)
NHERF2 - Na(+)/H(+) exchange regulatory cofactor NHE-RF2, PDZ domain (a.a. 147-230)
NHERF2 - Na(+)/H(+) exchange regulatory cofactor NHE-RF2, PDZ domain (a.a. 9-91)
NHERF4 - Na(+)/H(+) exchange regulatory cofactor NHE-RF4, PDZ domain (a.a. 326-419)
PDLIM1 - PDZ and LIM domain protein 1, PDZ domain (a.a. 1-87)
PDLIM2 - PDZ and LIM domain protein 2, PDZ domain (a.a. 1-82)
PDLIM4 - PDZ and LIM domain protein 4, PDZ domain (a.a. 1-87)
PDLIM5 - PDZ and LIM domain protein 5, PDZ domain (a.a. 1-85)
PDLIM7 - PDZ and LIM domain protein 7, PDZ domain (a.a. 1-84)
PDZK1 - Na(+)/H(+) exchange regulatory cofactor NHE-RF3, PDZ domain (a.a. 375-461)
PICK1 - PRKCA-binding protein, PDZ domain (a.a. 18-105)
RHPN2 - Rhophilin-2, PDZ domain (a.a. 514-598)
SCRIB - Protein scribble homolog, PDZ domain (a.a. 725-815)
SNTB2 - Beta-2-syntrophin, PDZ domain (a.a. 112-200)
SYNJ2BP - Synaptojanin 2 binding protein, PDZ domain (a.a. 5-101)
TAMALIN - GRP1 (general receptor for phosphoinositides 1)-associated scaffold protein, PDZ domain (a.a. 97-192)
TAX1BP3 - Tax1-binding protein 3, PDZ domain (a.a. 11-124)G-protein signaling
RGS1 - Regulator of G-protein signalling 1 (a.a. 63-205)
RGS10 - Regulator of G-protein signalling 10 [isoform b; a.a. 30-165)
RGS14 - Regulator of G-protein signalling 14 (a.a. 56-207)
RGS16 - Regulator of G-protein signalling 16 (a.a. 53-190)
RGS17 - Regulator of G-protein signalling 17 (a.a. 72-206)
RGS18 - Regulator of G-protein signalling 18 (a.a. 75-223)
RGS2 - Regulator of G-protein signalling 2, 24kDa (a.a. 71-203)
RGS3 - Regulator of G-protein signalling 3 [isoform 1], PDZ domain (a.a. 299-384)
RGS6 - Regulator of G-protein signalling 6 (a.a. 325-470)
RGS7 - Regulator of G-protein signalling 7 (a.a. 320-463)
RGS8 - Regulator of G-protein signalling 8 (a.a. 42-173)
Lipid signaling
ACOT11 - Acyl-coenzyme A thioesterase 11, START domain (a.a. 339-543)
ACOT12 - Acyl-coenzyme A thioesterase (a.a. 7-316)
ACOT4 - Peroxisomal succinyl-coenzyme A thioesterase (a.a. 1-421)
ACOT7 - Cytosolic acyl coenzyme A thioester hydrolase (a.a. 209-378)
bphA4 - Biphenyl dioxygenase ferredoxin reductase subunit (a.a. 1-408)
EPB41L3 - Band 4.1-like protein 3, FERM domain (a.a. 108-390)
FABP1 - Fatty acid-binding protein, liver (a.a. 1-127)
STARD13 - StAR-related lipid transfer protein 13, START domain (a.a. 903-1113)
STARD5 - StAR-related lipid transfer protein 5, START domain (a.a. 6-213)
Phosphoinositide dependent signaling - Lipid binding domains
IMPA2 - Inositol monophosphatase 2 (a.a. 16-288)
INPP5B - Type II inositol 1,4,5-trisphosphate 5-phosphatase (a.a. 339-643)
INPP5B - Type II inositol 1,4,5-trisphosphate 5-phosphatase (a.a. 341-646)
INPP5E - Phosphatidylinositol polyphosphate 5-phosphatase type IV (a.a. 275-623)
INPPL1 - Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 (a.a. 419-732)
NISCH - Nischarin, PX-domain (a.a. 18-124)
NUDT3 - Diphosphoinositol polyphosphate phosphohydrolase 1 (a.a. 1-172)
MIOX - Inositol oxygenase (a.a. 38-285)
SNX17 - Sorting nexin-17, PX domain (a.a. 1-108)
SNX7 - Sorting nexin-7, Pxdomain (a.a. 25-154)
TULP1 - Tubby-related protein 1, Tub family domain (a.a. 290-542)
Receptor signaling
G3BP1 Ras GTPase-activating protein-binding protein 1, NTF2 domain (a.a. 1-139)
SKIL Ski-like protein (a.a. 137-238)
TLR10 Toll-like receptor 10, TIR domain (a.a. 622-776)
Other signal
TTC5 - Tetratricopeptide repeat protein 5 (a.a. 261-424)
TWF2 - Twinfilin-2 (a.a. 6-137)
TWF2 - Twinfilin-2 (a.a. 179-313)
YWHAB - 14-3-3 protein beta/alpha (a.a. 1-240)
YWHAE - 14-3-3 protein epsilon (a.a. 1-233)
YWHAG - 14-3-3 protein gamma (a.a. 2-247)
YWHAH - 14-3-3 protein eta (a.a. 1-246)
YWHAQ - 14-3-3 protein theta (a.a. 1-234) - Amino acid metabolic enzymes, BTB-Kelch domain, hydrolases, transferases
Amino acid metabolic enzymes
AHCYL1 - S-adenosylhomocysteine hydrolase-like protein 1 (a.a 89-530)
AHCYL2 - Adenosylhomocysteinase 3, NAD binding domain (a.a. 175-607)
ASS1 - Argininosuccinate synthase P00966 (a.a. 1-412)
CSAD - Cysteine sulfinic acid decarboxylase (a.a. 1-493)
CTH - Cystathionine gamma-lyase (a.a. 1-402)
GFPT1 - Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, isomerase domain (a.a. 332-649)
GLUL - Glutamine synthetase (a.a. 5-365)
GPT2 - Alanine aminotransferase 2 Q8TD30 (a.a. 49-523)
MPST - 3-mercaptopyruvate sulfurtransferase (a.a. 11-289)
PCYT2 - Ethanolamine-phosphate cytidylyltransferase (a.a. 18-356)
PSAT1 Phosphoserine aminotransferase (a.a. 17-370)
SCLY - Selenocysteine lyase (a.a. 8-445)
TAT - Tyrosine aminotransferase (a.a. 41-444)
BTB-Kelch domain
BACH1 - Transcription regulator protein BACH1, BTB domain (a.a. 7-128)
BTBD6 - BTB/POZ domain-containing protein 6 (a.a. 129-236)
GAN - Gigaxonin, BTB domain (a.a. 6-126)
KLHL11 - Kelch-like protein 11 (a.a. 67-340)
KLHL12 - Kelch-like protein 12 (a.a. 267-567)
KLHL2 - Kelch-like protein 2 (a.a. 294-591)
KLHL7 - Kelch-like protein 7 (a.a. 283-586)
MYNN - Myoneurin, BTB domain (a.a. 4-117)
ZBTB33 - Transcriptional regulator Kaiso, BTB domain (a.a. 1-116)
ZBTB48 - Telomere zinc finger-associated protein, BTB domain (a.a. 4-120)Hydrolases
ADA - Adenosine deaminase (a.a. 5-363)
ECH1 - Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial, (a.a. 39-49,50-322)
ECHDC3 - Enoyl-CoA hydratase domain-containing protein 3, mitochondrial (a.a. 37-300)
ECHS1 - Enoyl-CoA hydratase, mitochondrial (a.a. 28-290)
ERAP1 - Endoplasmic reticulum aminopeptidase 1 (a.a. 1-505)
FH - Fumarate hydratase, mitochondrial (a.a. 44-510)
HDHD2 - Haloacid dehalogenase-like hydrolase domaing-containing 2 (a.a. 1-259)
HDHD3 - Haloacid dehalogenase-like hydrolase domain-containing 3 (a.a. 8-247)
HIBCH 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial (a.a. 32-386)
MDP1 - Magnesium-dependent phosphatase 1 (a.a. 1-165)
PTS 6-pyruvoyltetrahydropterin synthase (a.a. 7-145)
PUDP Pseudouridine-5'-phosphatase (a.a. 1-228)
Transferases
ACAA1 3-ketoacyl-CoA thiolase, peroxisomal (a.a. 30-424)
FDPS - Farnesyl pyrophosphate synthase (a.a. 67-419)
GGPS1 - Geranylgeranyl pyrophosphate synthase (a.a. 1-300)
GOT1 - Aspartate aminotransferase 1, cytoplasmic (a.a. 14-412)
HMGCS1 - Hydroxymethylglutaryl-CoA synthase, cytoplasmic (a.a. 16-470)
HMGCS2 - 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2 (mitochondrial)
MAT1A - Methionine adenosyltransferase I, alpha (a.a. 16-395)
MAT2A - Methionine adenosyltransferase II, alpha (a.a. 1-395)
MAT2B - methionine adenosyltransferase II, beta [RefSeq, isoform 2] (aka TGR, MAT-II, MGC12237, MATIIbeta, Nbla02999) (a.a. 28-334)
MCAT - Malonyl-CoA:acyl carrier protein transacylase (malonyltransferase) [isoform 1] (a.a. 60-375)
OXCT1 - 3-oxoacid CoA transferase (a.a. 40-520)
OXSM - 3-oxoacyl-[acyl-carrier-protein] synthase, mitochondrial (a.a. 39-459)
TKT - Transketolase (a.a. 10-620) - Miscellaneous ACACA - Acetyl-CoA carboxylase 1 (a.a. 78-617), Ligase
ACSM2A - Acyl-coenzyme A synthetase ACSM2A, mitochondrial (a.a. 32-577), Ligase AMP-Binding
ALDH1L1 - Cytosolic 10-formyltetrahydrofolate dehydrogenase (a.a. 1-307), Dehydrogenases & Reductases
ALOX12 - Polyunsaturated fatty acid lipoxygenase (a.a. 172-663) Arachidonic acid metabolism
AMPH – Amphiphysin (a.a. 34-236)
ARNT - Aryl hydrocarbon receptor nuclear translocator (a.a. 356-470)
BAK1 - Bcl-2 homologous antagonist/killer, mitochondrial apoptosis regulator (a.a. 21-190)
BCL2A1 - Bcl-2-related protein A1 (a.a. 1-149)
BIRC3 - Baculoviral IAP repeat-containing protein 3, BIR domain (a.a. 242-337), Apoptosis-Inflammation Domains
CA13 - Carbonic anhydrase 13 (a.a. 1-261), Lyase-Carbonic-Anhydrase
CA6 - Carbonic anhydrase (a.a. 20-290), Lyase-Carbonic-Anhydrase
CA7 - Carbonic anhydrase (a.a. 5-262), Lyase-Carbonic-Anhydrase
CA8 - Carbonic anhydrase-related protein (a.a. 1-290), Lyase-Carbonic-Anhydrase
CBLB - E3 ubiquitin-protein ligase CBL-B (a.a. 38-344), E3-Ligase
CBLC - E3 ubiquitin-protein ligase CBL-C (a.a. 9-323), E3-Ligase
CCNT2 - Cyclin-T2 (a.a. 7-263), Cyclin
CUL5 - Cullin-5, (a.a.) 1-386 ), E3-Ligase
DNPEP - Aspartyl aminopeptidase (a.a. 1-478), Protease
DPYSL2 - Dihydropyrimidinase-related protein 2 (a.a. 13-490), Dehydrogenases & Reductases
DPYS – Dihydropyrimidinase (a.a. 1-519), Dehydrogenases & Reductases
EBF1 - Transcription factor COE1, DNA binding domain (a.a. 10-250)
EBF1 - Transcription factor COE1, IPT/TIG domain (a.a. 258-351)
EBF3 - Transcription factor COE3, IPT/TIG domain (a.a. 261-395)
EBF3 - Transcription factor COE3, IPT/TIG domain, helix-loop-helix domain (a.a. 261-415)
ECI2 - Enoyl-CoA delta isomerase 2 (a.a. 137-394)
ELAC1 - Zinc phosphodiesterase ELAC protein 1, (a.a. 3-363), Metallo-Beta-Lactamase?
ENO3 - Beta-enolase (a.a. 1-434)
EPAS1 - Endothelial PAS domain-containing protein 1 (a.a. 239-350)
ERI3 - ERI1 exoribonuclease 3 (aa 137-337), Endo/Exo Nucleases
EXOG - Nuclease EXOG, mitochondrial (a.a. 42-368), Endo/Exo Nucleases
GHRHR - Growth hormone-releasing hormone receptor (a.a. 34-123), Membrane-GPCR-ECD
GLS - Glutaminase kidney isoform, mitochondrial (a.a. 221-533)
GUCY1B1 - Guanylate cyclase 1, soluble, beta-1 (a.a. 408-619), Cyclase-Nucleotide
GYG1 - Glycogenin 1 (a.a. 1-262)HBP1 - HMG-box transcription factor 1 (a.a. 206-342)
HSP90B1 - Heat shock protein HSP 90-beta (a.a. 284-543)
HTD2 - Hydroxyacyl-thioester dehydratase type 2, mitochondrial (a.a. 23-168), Lyase-Other
LASP1 - LIM and SH3 domain protein 1, SH3 domain (a.a. 202-261)
LCN15 - Lipocalin-15 (a.a. 19-177), Lipocalin
MACROD1 - ADP-ribose glycohydrolase MACROD1, MACRO domain (a.a. 91-325), Deacetyalases
MACROH2A2 - Core histone macro-H2A.2 (a.a. 177-369)
MAGEA4 - Melanoma antigen family A, 4 (a.a. 99-317), MAGE
MLYCD - Malonyl-CoA decarboxylase (a.a. 40-490), Lyase-Other
MMACHC - Cyanocobalamin reductase or alkylcobalamin dealkylase (a.a. 1-282)
MMUT - Methylmalonyl-CoA mutase, mitochondrial (a.a. 12-750)
NAIP - Baculoviral IAP repeat-containing protein 1, BIR2 domain (a.a. 139-244), Apoptosis-Inflammation Domains
NT5C2 - Cytosolic purine 5'-nucleotidase (a.a. 1-536)
NT5C3 Cytosolic 5'-nucleotidase 3A (a.a. 64-336)
NUDT - Oxidized purine nucleoside triphosphate hydrolase (a.a. 42-197)
NUDT21 - Cleavage and polyadenylation specificity factor subunit 5 (21-227), RNA processing
NUMBL - Numb-like protein (a.a. 60-204, Neuronal stem cell development
PCCA - Propionyl-Coenzyme A carboxylase, alpha polypeptide precursor (a.a. 658-728), Ligase
PTGDS - Prostaglandin-H2 D-isomerase (a.a. 23-190), Arachidonic acid metabolism
PMP2 - Myelin P2 protein (a.a. 3-132), Lipocalin
RADIL - Ras-associating and dilute domain-containing protein, Ras association domain (a.a. 51-193), Cell-type specific expression - endothelial cell
RAMP2 - Receptor activity-modifying protein 2 (a.a. 48-139), Membrane-GPCR
RECQL - ATP-dependent DNA helicase (a.a. 49-616), DEAD-DNA-Helicase
SOCS6 - Suppressor of cytokine signaling 6, SH2 domain (a.a. 3611-499), SOCS-box-containing protein
SPSB1 - SPRY domain-containing SOCS box protein 1 (a.a. 24-233), SOCS-box-containing protein
SPSB2 - SPRY domain-containing SOCS box protein 2 (a.a. 26-219), SOCS-box-containing protein
SPSB4 - SPRY domain-containing SOCS box protein 4 (a.a. 28-233), SOCS-box-containing protein
SYN3 - Synapsin 3 (a.a. 76-417), Vesicle-Transport
TATDN1 - Deoxyribonuclease TATDN1 (a.a. 5-295)
TATDN3 - Putative deoxyribonuclease TATDN3 (a.a. 5-274)
TGM1 - Protein-glutamine gamma-glutamyltransferase K, Ig domain (a.a. 693-787), Transglutaminase
TH - Tyrosine 3-monooxygenase (a.a. 193-528), Neurotransmitter-Synthesis
TP73 - Tumor protein p73 (a.a. 112-311)
VIPR2 - Vasoactive intestinal polypeptide receptor 2, hormone receptor domain (a.a. 26-118), Membrane-GPCR-ECD
ZFYVE16 - Zinc finger FYVE domain-containing protein 16, FYVE zinc finger domain (a.a. 733-820)
Gene to structure services
SARomics Biostructures' X-ray crystallography services platform also provides gene-to-structure services, which include cloning, expression, purification of proteins, and characterization using biophysical methods, followed by crystallization, X-ray data collection either at the MAX IV synchrotron in Lund or at some other European synchrotron, followed by high resolution structure analysis. For optimal handling, high-throughput protein characterization and crystallization protocols have been developed at the company and used throughout the process.
Gene-to-Structure services are performed on a fee-for-service basis and include:
More details on the methods used in X-ray crystallography services can be found on our Learning Center protein crystallization page. Contact us directly to discuss your project using the contact form.
Gene-to-Structure services are performed on a fee-for-service basis and include:
- Cloning, expression, and purification of the protein
- Biophysical characterization using dynamic light scattering (DLS), CD spectroscopy, differential scanning fluorimetry (DSF), and, if appropriate, NMR spectroscopy.
- Crystallization, X-ray data collection, and structure determination.
- Protein-ligand co-crystallization and structure determination.
More details on the methods used in X-ray crystallography services can be found on our Learning Center protein crystallization page. Contact us directly to discuss your project using the contact form.

Maria Håkansson, PhD, Principal Scientist, Team Leader, Protein Crystallography, and Raymond Kimbung, PhD, Senior Scientist, Team Leader, Protein Production
Crystallography services outline: Cloning, expression, purification and characterization
Obtaining well-diffracting crystals is an art. To accelerate the process, we have developed proprietary protocols for the optimal handling of all project steps. The first step is defining the correct protein construct, followed by cloning, expression, and purification. Construct definition is essential since, in many examples removing just a couple of amino acid residues, e.g., at the N-terminus, dramatically improved protein crystal quality and the quality of the X-ray data. Among other methods, NMR spectroscopy can verify if the construct yields a folded, well-behaved protein. In addition, other biophysical methods like Dynamic Light Scattering (DLS), CD spectroscopy, and Differential Scanning Fluorimetry (DSF, also called thermal shift assay) may be used for verification. Obtaining a monodisperse solution of the protein is essential for successful crystallization. For illustration, a product sheet that shows the characteristics of a crystallization-grade protein from our Protein Shop can be downloaded here.

A plate hotel is used for storing and monitoring crystallization plates, usually kept at a constant temperature.
The 96-well crystallization plate from Hampton Research.
The 96-well crystallization plate from Hampton Research.
Crystallization & synchrotron X-ray data collection
X-ray protein crystallography is central to structural biology. And protein crystallography requires crystals! If crystallization conditions are unknown, we use our commercial and proprietary screens collection to identify initial crystallization conditions. Crystallization robotics is used in this process, while the crystallization plates are kept in plate hotels at a constant temperature. The plates are periodically monitored using an automated imaging system. Usually, the initial conditions need to be optimized to obtain well-diffracting crystals. The crystals are tested using synchrotron radiation. On average, we collect X-ray data on European synchrotrons every second week, on-site or remotely. Lund's MAX IV synchrotron radiation facility is just a few kilometers from our lab. Please see our technology pages for additional details on our protein X-ray crystallography services.
X-ray structure determination
Model building and refinement can start after collecting good-quality X-ray data and obtaining an electron density map. At this stage, the presence of a ligand (in the case of a protein-ligand complex) can quickly be assessed. Our customers are periodically informed of the project's progress, and any deviations from the plan are discussed before proceeding.
More details on sample preparation can be found in our guidelines for preparing and sending samples for crystallization.
Please do not hesitate to contact us if you require assistance preparing samples or any other information.
More details on sample preparation can be found in our guidelines for preparing and sending samples for crystallization.
Please do not hesitate to contact us if you require assistance preparing samples or any other information.
Recent publications
Zetterberg FR, Diehl C, Håkansson M, Kahl-Knutson B, Leffler H, Nilsson UJ, Peterson K, Roper JA & Slack RJ (2023).
Discovery of Selective and Orally Available Galectin-1 Inhibitors
J. Med. Chem. https://doi.org/10.1021/acs.jmedchem.3c01787
PDB entries:
8OJP - Human galectin 1 in complex with inhibitor (on hold until publication)
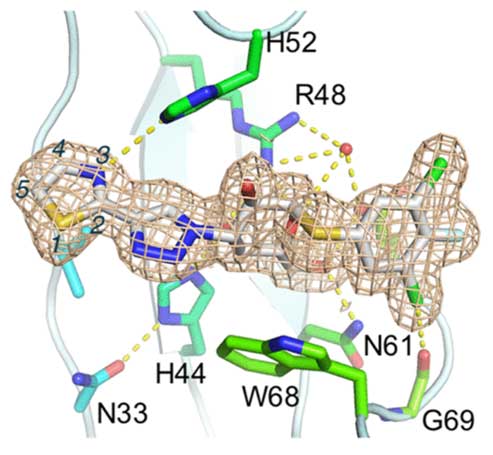
A new series of orally available α-d-galactopyranosides with high affinity and specificity toward galectin-1 have been discovered. High affinity and specificity were achieved by changing six-membered aryl-triazolyl substituents in a series of recently published galectin-3-selective α-d-thiogalactosides (e.g., GB1107 Kd galectin-1/3 3.7/0.037 μM) for five-membered heterocycles such as thiazoles. One compound, GB1490 (Kd galectin-1/3 0.4/2.7 μM), was selected for further characterization toward a panel of galectins showing a selectivity of 6- to 320-fold dependent on galectin. The X-ray structure of GB1490 bound to galectin-1 reveals the compound bound in a single conformation in the carbohydrate binding site.
For other publications please see our publications page
Discovery of Selective and Orally Available Galectin-1 Inhibitors
J. Med. Chem. https://doi.org/10.1021/acs.jmedchem.3c01787
PDB entries:
8OJP - Human galectin 1 in complex with inhibitor (on hold until publication)
A new series of orally available α-d-galactopyranosides with high affinity and specificity toward galectin-1 have been discovered. High affinity and specificity were achieved by changing six-membered aryl-triazolyl substituents in a series of recently published galectin-3-selective α-d-thiogalactosides (e.g., GB1107 Kd galectin-1/3 3.7/0.037 μM) for five-membered heterocycles such as thiazoles. One compound, GB1490 (Kd galectin-1/3 0.4/2.7 μM), was selected for further characterization toward a panel of galectins showing a selectivity of 6- to 320-fold dependent on galectin. The X-ray structure of GB1490 bound to galectin-1 reveals the compound bound in a single conformation in the carbohydrate binding site.
For other publications please see our publications page