Compound library screening in drug discovery aims to identify a compound or a series of compounds that can bind to the drug target and modulate its activity. Often, this process is compared to finding a needle in a haystack. However, it does not need to be this way. The process can be made much more reliable and predictable with the right choice of compound library and the screening method. In an earlier blog post, we discussed the basic principles of library generation and the various experimental methods used in screening. Here, we continue the theme and discuss weak affinity chromatography (WAC™), the primary fragment screening method used within our lead discovery services program. It is a proprietary technique jointly owned by SARomics Biostructures and our partner, Red Glead Discovery.
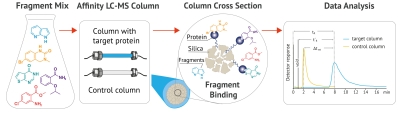
WAC™ is a version of affinity chromatography that detects weak-affinity ligand binding to a target molecule. In contrast to traditional affinity chromatography, where the molecule of interest is tightly bound to the immobilized target and subsequently eluted e.g. with drastic pH or an ionic strength shift, the WAC™ technology offers a gentle approach. A mixture of small molecular weight fragments or larger compounds is injected into a standard high-performance liquid chromatography (HPLC) column packed with silica on which the protein target has been immobilized. During elution, the fragments with a higher affinity for the protein will stay on the column longer than those with a low affinity. Although the binding of small fragments is usually very weak, with Kd often in the mM range, modern methods such as mass- or UV spectrometry allow for the efficient detection and classification of the binders. With the ability of simultaneous identification of weak binders (mM) in complex mixtures, WAC™ has proven to be an efficient and low-cost choice compared to other fragment screening methods, e.g., surface plasmon resonance (SPR), X-ray crystallography, NMR spectroscopy, affinity selection-mass spectrometry (AS-MS) or thermal shift assay (TSA).
The WAC™ fragment screening method originates from the academic research of Prof. Sten Ohlson and his co-workers. Its advantages have been validated in over 50 projects we have run on around 20 distinct target classes over the past seven years since its commercial implementation. Among the advantages we can highlight:
- Physiological buffer as a mobile phase
- Label-free and detection by MS (enables built-in quality control)
- Straightforward detection of retention times from which Kd can be calculated
- Potential for high throughput screening (up to 5000 fragments per week)
- Low material consumption (<5 mg protein)
- mM hits can be identified by screening fragments at low concentrations (1-5 µM)
- Immediate hit ranking and control of hit quality
- High hit rates from 1-20% (avg 6%)
WAC™-projects run by SARomics Biostructures & Red Glead Discovery included screening for novel fragment hits as project starting points, assessing the druggability of new targets, ranking of fragment hits identified by X-ray crystallographic fragment screening (XFS), finding differentiated backups in mature projects, and as a rescuing mode for challenging targets (PPIs, etc.). The target classes included kinases, GTPases, cytokine/protein-protein interactions (PPI), bromodomains, integrins, helicases/ATPases, transcription factors, deubiquitinases, ubiquitin ligases, and many others.
After initial hit identification, follow-up activities may include fragment hit expansion by accessing our integrated lead discovery platform built around our joint capabilities in medicinal chemistry, computational chemistry, X-ray crystallography, biophysical validation, biochemical and cell-based assays, in vitro ADME, etc. Protein NMR spectroscopy and X-ray crystallography can be used to verify compound binding and to gain insights into the details of the interactions with the target protein. In addition, the WAC™ column lasts for a few to several months and can be used repeated times for fast ranking of acquired or synthesized analogs.
Case studies
A recent case study of WAC™ fragment screening of interleukin-23 (IL-23) run by the SARomics Biostructures & Red Glead Discovery team was reported at the Drug Discovery Chemistry conference in San Diego (April 2024). IL-23 belongs to a class of inflammatory proteins called cytokines involved in signal transduction between cells. Monoclonal antibody inhibitors of IL-23 that block the action of IL-23 have been approved for clinical treatment of inflammation, and an orally administered small-molecule inhibitor of the IL-23 pathway has the potential to provide significant benefits for patients suffering from autoimmune inflammatory disorders. The reported case study used a library of 1013 fragments for the WAC™ screening. With a hit rate of 2%, 19 fragments were identified, of which six were nominated for co-crystallization with the target. Two complex structures at resolutions of 2.7 Å and 2.95 Å were obtained. Although the two fragments do not block biologically relevant receptor interactions, the analysis of their binding mode provides valuable insights for further expansion of the molecules towards the design of efficient inhibitors.
Another case study involved screening of the transcription regulator BRG1 (SMARCA4). This protein is part of the large chromatin remodeling complex SWI/SNF, involved in transcriptional gene activation. A ligandability screen with 250 compounds gave 20 hits. Five crystal structures of complexes with ligands were solved, and a new ligand binding site was identified (for details, see poster).
WAC™ fragment screening offer
We are currently unlocking the power of weak affinity chromatography for a broader audience with our new, more affordable package deals! We're excited to introduce more accessible options, and one of our latest fixed-price offers features a comprehensive WAC™ screen of 500 fragments, coupled with analog search, selection, and WAC testing. WAC™ is also ideal for ranking your weak affinity hits from XFS. Please contact us to know more!