Structural Biology of Therapeutic Monoclonal Antibodies
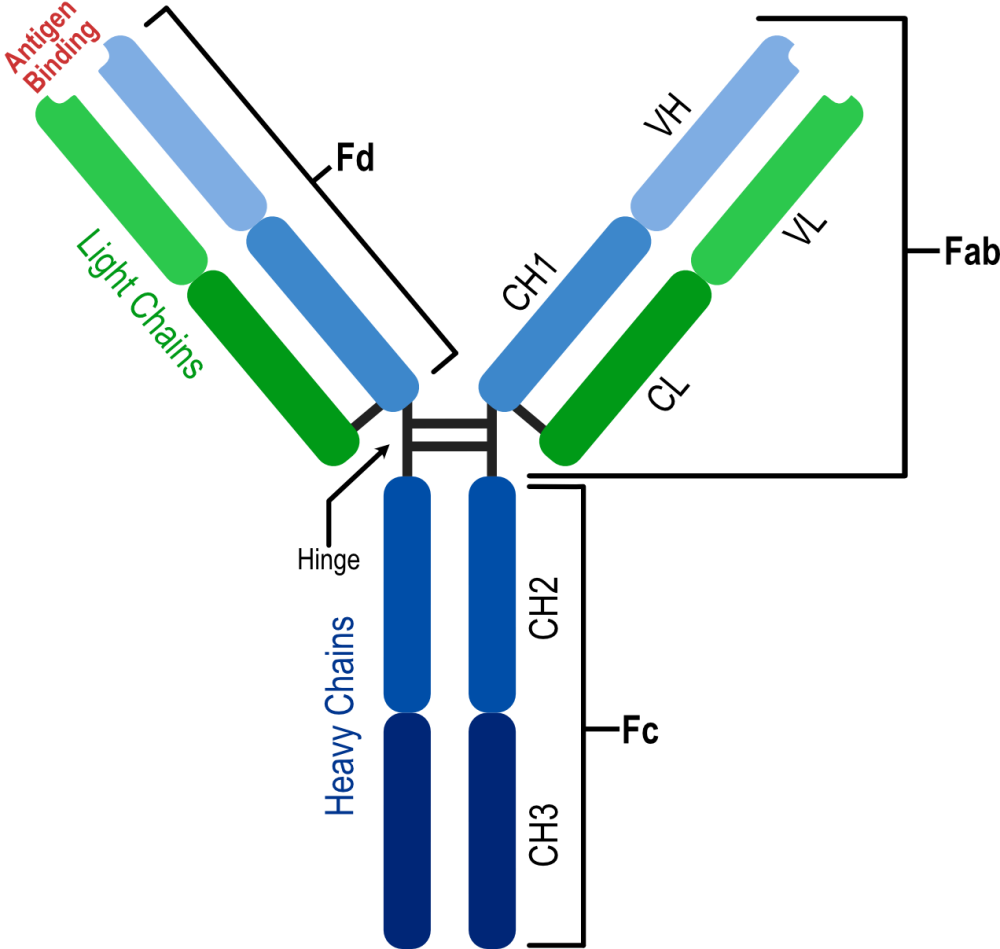
X-ray crystallography has provided detailed information on the three-dimensional structure of monoclonal antibodies, revealing domain organization, dynamics, and the structure of the paratope and epitope regions. Image above was generated by Affinity Designer 2, https://geblerconsulting.com/
The structural biology of monoclonal antibodies has greatly advanced our understanding of the mechanisms of their function. This has also resulted in many experimentally determined antibody three-dimensional structures. Thus, the Structural Antibody Database (SabDab) now contains a staggering 7471 antibody structures and 7151 structures of antibody-antigen complexes as of July 2023. Through X-ray crystallography and the collaborative efforts of numerous research groups, structural biology has provided a comprehensive explanation of the fundamental aspects of antibody structure and dynamics, offering valuable insights into how antibody structure relates to its function.
The structural biology of monoclonal antibodies and antibody-antigen complexes has focused on detailed studies of the paratope (the antibody residues contacting the antigen) and epitope (the antigen residues involved in stabilizing the antibody-antigen complex), domain organization and dynamics, including flexibility of the different parts of the structure. It has helped to delineate the parts of the antibody structure responsible for antigen binding – the complementarity-determining region loops (CDRs) and the supporting framework regions (FRs), which often participate directly in antigen binding.
The development of new therapies using biologics, the majority of which are monoclonal antibodies, is a significant focus of modern-day drug design. The information on antibody three-dimensional structure, combined with antibody engineering, plays a central role in this process and has opened the way for the rapid growth of monoclonal antibody therapeutics. As of June 2023, the Antibody Therapies Database (https://umabs.com/) contains information on over 9400 monoclonal antibodies targeting over 2400 human disorders ranging from cancer and rheumatoid arthritis to osteoporosis and asthma. Antibodies are considered attractive therapeutic molecules due to their specificity, long half-life in vivo (two to four weeks), and lack of immunogenicity for fully humanized antibodies.
The structural biology of monoclonal antibodies has also helped design new antibody variants suitable for diagnostic purposes (such as immunofluorescence, Western blotting, and enzyme-linked immunosorbent assay (ELISA) analysis). As in the case of small-molecule drug discovery, structural biology, and structure-based antibody engineering play a crucial role, both at the early stages of design and development and at the later stages of clinical studies. Structural information, combined with monoclonal antibody engineering, can help optimize the chemical properties of antibodies, like selectivity, solubility, and efficacy, and can even help overcome drug-resistant mutations. Structural information can also be important in patent applications and intellectual property protection. Structure-guided design of new mutations in developing new therapeutic monoclonal antibodies may also improve their stability and resistance against degradation and aggregation in blood and during production, storage, and transportation.
Humanization
Developing new monoclonal antibodies against human targets usually starts with non-human antibodies from rodents, chickens, or rabbits. These antibodies require humanization. The availability of the three-dimensional structure of the antibody-antigen complex or even just the antibody greatly facilitates the definition of the CDR and framework regions, revealing the residues involved in complex stabilization. Humanization proceeds in several steps, first defining the CDRs of the non-human antibody, after which the human sequence to be utilized as the donor of the rest of the antibody needs to be selected. Some residues of the framework region and the CDRs must be replaced to reduce immunogenicity while maintaining the engineered molecule’s activity. The three-dimensional structure helps design these replacements by so-called back mutations (replacing non-human amino acids) and “resurfacing”.
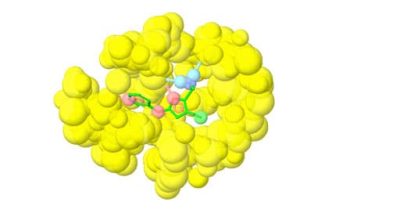
The three-dimensional structure of the antibody-antigen complex can also be used to design bispecific monoclonal antibodies. In a recent publication by Beckmann et al., the development of a platform of dual targeting Fab (DutaFab) molecules, which comprise two spatially separated and independent binding sites within the human antibody CDR loops, was reported. The CDRs were separated into the H-side paratope containing loops HCDR1, HCDR3, and LCDR2 and the L-side paratope containing loops LCDR1, LCDR3, and HCDR2. The SARomics Biostructures team crystallized the complex with the antigens and determined the X-ray structures (shown in the image above). The structures showed that DutaFabs can simultaneously bind two target molecules at the same Fv region comprising a VH-VL heterodimer of the Fab.
The SARomics team has gathered extensive experience in monoclonal antibody and antibody-antigen complex crystallization and structure determination. Our customers have published many structures contributed by our team. Many examples can be found on our antibody structure services and publications pages.
The article is our first introductory post dedicated to antibody therapeutics. The second post is focused on the exciting area of biosimilars, the design of which is also helped by structural information, and the third post is on the advantage of NMR spectroscopy in biosimilar comparability analysis. Follow us on LinkedIn to ensure you don’t miss our future posts.
Reference
Beckmann R, Jensen K, Fenn S, Speck J, Krause K, Meier A, Röth M, Fauser S, Kimbung R, Logan DT, Steegmaier M & Kettenberger H (2021). DutaFabs: A novel platform of bispecific therapeutic Fab fragments that simultaneously bind two targets with high affinity. Nature Communications 12, 708. https://doi.org/10.1038/s41467-021-20949-3
The image above was generated by Affinity Designer 2.
For additional information on the subject, please see the excellent review below:
Chiu ML, Goulet DR, Teplyakov A, and Gilliland GL (2019). Antibody structure and function: the basis for engineering therapeutics. Antibodies, 8(4), 55; https://doi.org/10.3390/antib8040055